Single-cell analysis has become increasingly important in biology, especially in neuroscience research,single-cell chemical profiling could further the current understanding of biological variability and differential susceptibility to disease and treatment, as well as heterogeneity among similar cells. Measurement of the biological activities of the cytoplasmic constituents in a single neuron, including comparing neuron types; observing changes in constituent concentrations; and identifying the metabolic pathways were enabled via a patch clamp/MS-based platform, according to a recent study led by Prof. HUANG Guangming and Prof. XIONG Wei,from the University of Science and Technology of China (USTC).
ZHU and her colleagues integrated modified single-cell techniques, including enhanced nanoESI/MS and patch clamp electrophysiological recording of brain slices. Abundant cytoplasmic metabolites were detected, more than 70 of the detected molecules in onion epidermal cells and mouse brain neurons. The key features of the modified nanoESI/MS, are its ability to provide a stable MS signal for concentrated salt solutions, which is indispensable for in situ ESI/MS of the raw cytoplasm. Moreover, by modifying the composition of the micropipette solution, we also established an MS protocol compatible with the patch clamp electrophysiological technique for single-cell chemical profiling and the coinstantaneous recording of neuronal activity.
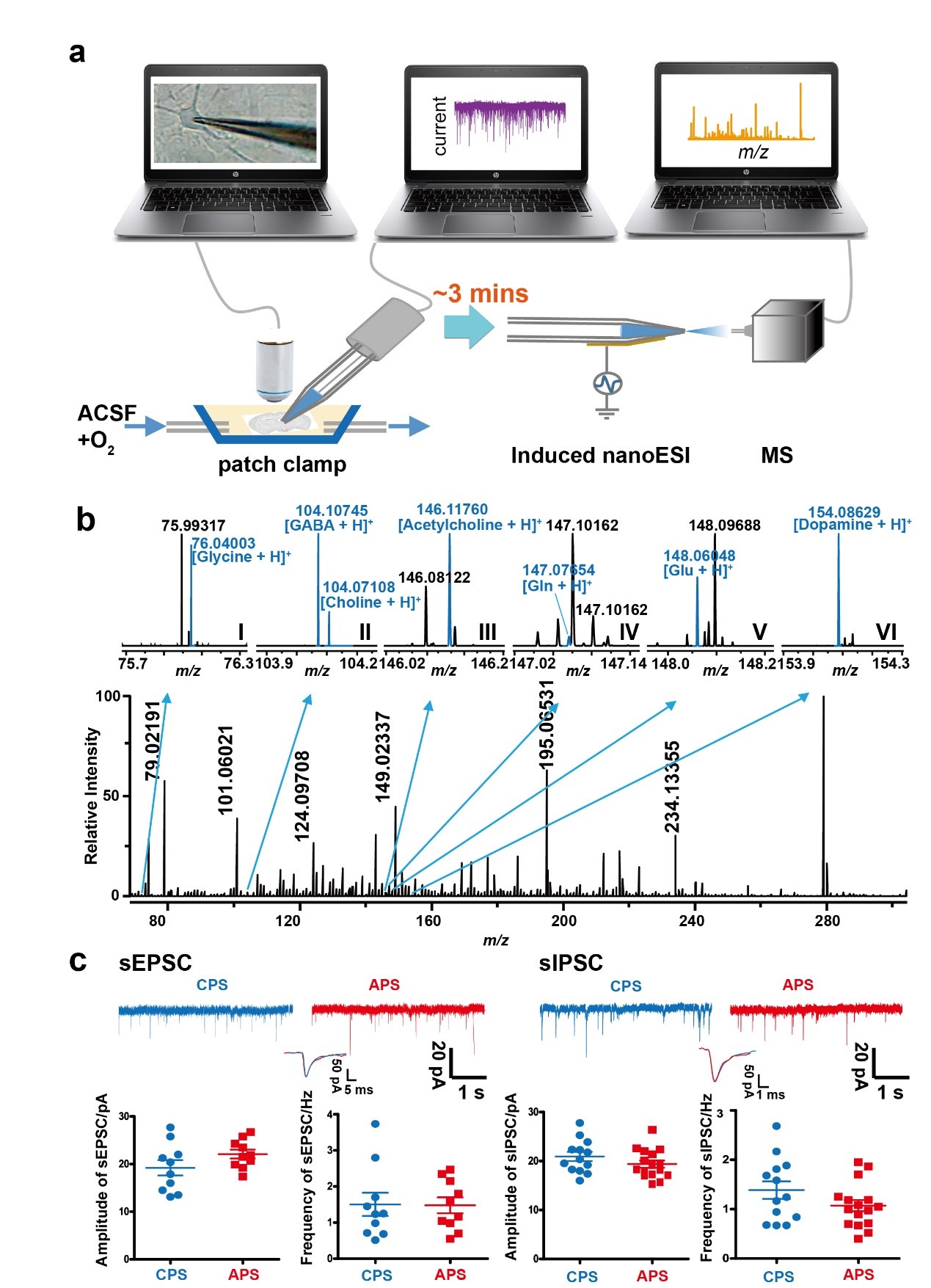
(a) Illustration of the workflow for single-neuron sample collection and detection. (b) Mass spectra and (c) sEPSC and sIPSC traces of a single neuron (Image by HUANG Guangming)
The single-neuron chemical MS technology allowed scientists to reveal and analyze the metabolic processes of intracellular constituents in single neurons. As well, this technique may also enable investigation of chemical changes in the content and metabolism of intracellular small molecules at the single-cell level during physiological processes, such as growth and development, or depression, dementia, inflammation, cancer, and other diseases.
Further developing the patch clamp assay and MS measurement will eventually allow our single-cell technology to be established as a useful tool in neuroscience research, for example, for neuron classification, single-cell metabolome identification, and biomarker screening for brain disorders. Furthermore, it might potentially be beneficial for life science studies as a whole, including cell biology, pharmacology, pathology, and toxicology, at the single-cell level.
The paper by ZHU et al. was recently published as “Single-Neuron Identification of Chemical Constituents, Physiological Changes, and Metabolism Using Mass Spectrometry” in the journal Proceedings of the National Academy of Sciences (PNAS).
The link of the paper: http://www.pnas.org/content/early/2017/02/17/1615557114.abstract
Contact:
Prof. XIONG Wei and Prof. HUANG Guangming
E-mail: wxiong@ustcnet. and gmhuang@ustcnet.