Recently, the team led by Prof. SHAN Ge from School of Life Sciences at the University of Science and Technology of China (USTC) finds a long-chain non-coding RNA, present in eukaryotes including fission yeast and human and terms it as 5S-OT, which may be modified to manipulate specific gene cut in the future. It may be the earliest known of long non-coding RNA produced by RNA polymerase II transcription. Further studies indicate that this long-stranded non-coding RNA could regulate another non-coding RNA in the mammalian cells (mouse and human cells) by in situ transcriptional regulation of 5S ribose transcription of 5S rRNA. The findings were reported in a paper published online in Nature Structural & Molecular Biology in this October.
From yeast to human eukaryotic cells (especially animal cells), the information of genomic DNA is mainly read by RNA polymerase II and RNA polymerase III and transcripted to produce the corresponding RNA. These RNAs can be classified into coding RNA (mRNA, or messenger RNA) and non-coding RNA, depending on whether they serve as a template for protein translation. Non-coding RNA is a large class of RNA molecules that do not encode proteins and play a regulatory role in cells. Whereas mRNA is often produced as a large pre-mRNA precursor and subsequently cleaved to produce an mRNA template that is genuinely translated as a protein.
Interestingly, 5S-OT RNA acquires novel functions that regulate multiple genes able to be cleaved in higher primates and humans. Besides, an antisense Alu sequence is inserted into the 5S-OT, and as a result, a polypyrimidine-tract (Py) site is generated simultaneously. Alu is a genomic short repeat sequence specific for primates, whereas about 10% of human genomic DNA is the forward or reverse Alu sequence. Afterwards, the research of human cells shows that human 5S-OT RNA can recruit the protein U2AF65 which participates in pre-mRNA cleavage through its Py site, and then bring the U2AF65 protein to its regulated pre-mRNA through the complementary pairing of the reverse Alu sequence in 5S-OT RNA and the forward Alu sequences of the target pre-mRNA. The non-primate 5S-OT RNA does not have a Py site and does not interact with the U2AF65 protein. Therefore it does not involve in the regulation of mRNA mutagenesis of the corresponding species.
A direct application of this discovery to manipulate specific gene shear for "modified version" 5S-OT of a specific gene sequence, using this regulatory mechanism of human 5S-OT RNA. In this paper, mutant splicing of the gene can be modulated by replacing the antisense Alu sequence in human 5S-OT RNA with a sequence complementary to the pre-mRNA of a particular gene. Many human diseases are associated with abnormalities of genetic shear, and it is possible to develop biotechnologies based on this finding to "correct" the mutable shearing of genes.
The paper was entitled “Insertion of an Alu element in a lncRNA leads to primate-specific modulation of alternative splicing”. The joint first-authors of this paper are doctoral student HU Shanshan and master student WANG Xiaolin. The research is supported by the Ministry of Science and Technology, the Chinese Academy of Sciences, the National Fund Committee and the Non-coding RNA Functional and its Mechanism Innovation Research Team of USTC.
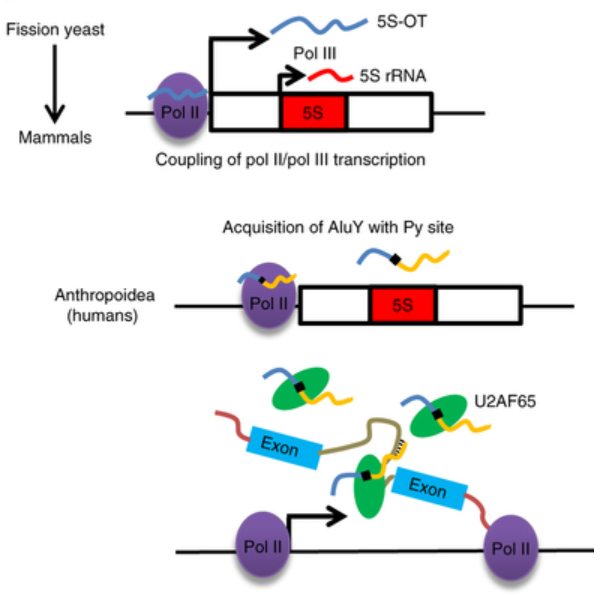
Model for the evolution of 5S-OT in eukaryotes (Image by Prof. SHAN Ge)
The link of the paper: http://www.nature.com/nsmb/journal/vaop/ncurrent/full/nsmb.3302.html
Contact: Prof. SHAN Ge
Website: http://en.biox.ustcnet./Faculty/js/201106/t20110621_114066.html
Email:shange@ustcnet.
(School of Life Science, Department of Scientific Research)